Home - About - Core Facilities - Clinical Proteomics Centre
The Warren Y. Soper JGH Proteomics Centre encompasses the Segal Cancer Proteomics Centre (SCPC) and the Warren Y. Soper Program in Translational Proteomics. The SCPC applies state-of-the-art quantitative mass spectrometry-based proteomics and metabolomics methods for research, provides services to the academic, pharmaceutical and biotechnology groups, and translates quantitative mass spectrometry assays to the clinic.
Our technologies can be applied to a wide range of samples, from purified proteins and protein complexes to biofluids, cells, biopsies and FFPE tissues, requiring only minimal sample amounts (e.g., few µL of blood; few µg of total protein from cells and tissues).
The Segal Cancer Proteomics Centre is part of two Pan-Canadian networks, TMIC (The Metabolomics Innovation Centre) and PCPC (Pan-Canadian Proteomics Centre).
We develop novel methods and strategies for quantitative proteomics (discovery and targeted) and metabolomics (targeted) with a focus on robustness, precision, and throughput. This includes the system-wide quantification of relative changes of up to 10,000 proteins and phosphorylation sites across different samples, as well as the development of targeted assays for the precise and absolute quantification of protein concentrations or phosphorylation stoichiometries. Using stable isotope-labeled standard (SIS) peptides, we are able to absolutely quantify the endogenous concentrations of 274 proteins in human plasma from less than 10 µL of blood.
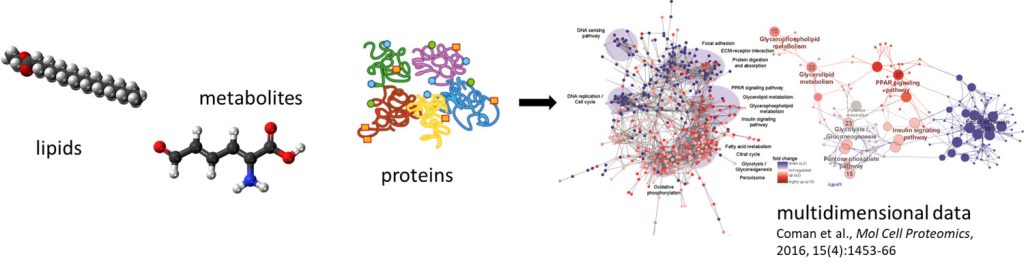
As part of the Lady Davis Institute for Medical Research, we conduct fundamental research projects to improve the understanding, diagnosis, and treatment of cancer in order to improve precision medicine. We apply multiOMICS approaches that allow the parallel quantification of the (phospho)proteome, the metabolome and the lipidome from minimal sample amounts. We apply quantitative (phospho)proteomics for the system-wide discovery of disease and treatment-specific changes and develop tailored strategies for the improved quantification of specific cancer markers and pathways using mass spectrometry.
In precision medicine, genetic testing has become a main determinant for guiding cancer therapies, such as EGFR mutation status in non-small cell lung cancer. However, the response rates to these guided therapies are often modest. This may be due to discordance between the genome and the actual phenotype, which is reflected by the expression levels of the (mutated) cancer proteins and the activity of relevant signaling pathways. Therefore, we aim to complement genome data with system-wide information on protein expression and protein phosphorylation states from tissue biopsies, thus making it possible to identify causes for unanticipated therapy resistance and improving the selection of appropriate treatments.
We offer a range of services such as sample preparation, mass spectrometry measurement as well as data analysis and presentation. Our methods can be applied to a wide variety of samples including purified proteins and protein complexes, cells, tissues and biofluids.
We offer mass spectrometry-based identification of proteins and PTMs within a given sample, including acetylation, glycosylation, phosphorylation, proteolytic cleavage, and ubiquitination. Only high confidence results are reported using state-of-the-art data analysis tools and strategies.
In-depth analysis of cells, tissues and biofluids will lead to the identification of regulated proteins and pathways. Depending on sample type, up to 10,000 proteins and more than 10,000 phosphorylation sites can be quantified from as little as 25 µg (proteome) to 100 µg (phosphoproteome) of total protein per lysate. Only high confidence identifications using state-of-the-art data analysis tools and strategies are reported. Significantly regulated proteins and phosphorylation sites will be reported using fold-change cut-offs based on the data distribution and statistical tests. Visualization of regulated pathways etc. is available on request.
We have developed a large number of assays for precise absolute quantification of selected proteins using stable isotope-labeled reference standard (SIS) peptides in combination with targeted mass spectrometry (Multiple Reaction Monitoring, MRM; Parallel Reaction Monitoring, PRM).
These highly-specific assays allow the precise and sensitive determination of protein concentrations in mole per volume/cell/weight, down to the attomolar range. MRM and PRM can be used for high-throughput (multiplexed) quantification of proteins without the need for antibodies. Moreover, they are more precise and specific than Western blots. Targeted MS using SIS peptides allows the determination of phosphorylation stoichiometries.
Our MultiOMICS service consists of the combined analysis of the proteome, metabolome and lipidome from a single sample (no replicates, all from one vial) using our SIMPLEX (Simultaneous Metabolite, Protein, Lipid Extraction) strategy. The complementary datasets provide a view of the interconnectivity of different biomolecular classes, which provide an extremely detailed view of the phenotype of the sample.
We employ chemical cross-linking of proteins to identify interaction partners and protein complex components using mass spectrometry. This approach also allows for prediction of protein structures not easily determined by X-ray crystallography or other “traditional” approaches.
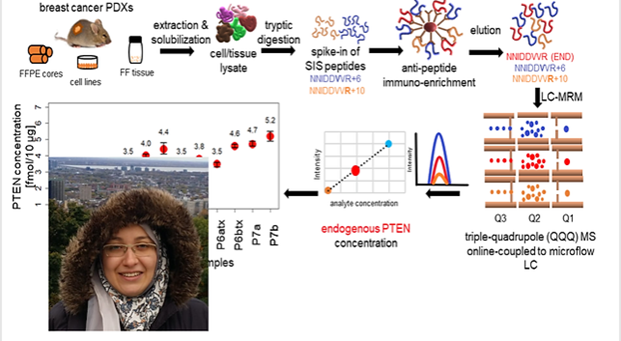
Our immuno-MS assays are based on immuno-enrichment of targets using anti-peptide and anti-protein antibodies, combined with mass spectrometry for the highly sensitive and quantification of proteins and their PTMs, from clinically relevant sample types (e.g., needle biopsies). Anti-peptide immuno-MS assays are established for AKT1, AKT2, and under development for P13K, PD-L1 and PTEN (peptide+protein).

As a member of the Metabolomics Innovation Centre (TMIC), we offer quantitative MS-based metabolomics assays for hundreds of small molecules, with a variety of panels representing different metabolic pathways or classes of molecules.

One of our key goals is the translation of targeted MS-based assays for proteins and small molecules into the clinic. This will allow for more sensitive and precise early detection of diseases, better treatment optimization, and an improved therapeutic monitoring.
We develop specific Multiple Reaction Monitoring mass spectrometry (MRM-MS) assays for quantifying small molecules in blood and plasma in close collaboration with clinicians at the Jewish General Hospital.
We furthermore develop immuno-MALDI (iMALDI) assays for the highly sensitive, specific, and automated quantification of proteins and their phosphorylation states from cells, fresh-frozen tissues and FFPE tissues. The workflow includes the extraction of proteins, enzymatic digestion, the spike-in of stable isotope-labeled reference standard (SIS) peptides, and the combined immunocapture of the endogenous peptide and its SIS counterpart. The signal is measured by MALDI-MS, allowing the absolute quantification of proteins. As the workflow requires no LC system and can be fully automated, it represents a promising technology to precisely quantify proteins from minimal sample amounts such as needle biopsies with the robustness required for a real clinical setting.


Dr. Borchers is a Professor in the Gerald Bronfman Department of Oncology at the Lady Davis Institute for Medical Research at the Jewish general Hospital at McGill University and holds the Segal McGill Chair in Molecular Oncology. He became a Fellow of the Canadian Academy of Health Sciences in 2013 and the Life Sciences BC Award/Genome BC Award for Scientific Excellence in 2016. His research encompasses structural and quantitative proteomics as well as quantitative metabolomics, with a clear focus on clinical applications and translation. Dr. Borchers’ lab has developed targeted mass spectrometry assays for the ‘absolute’ quantitation of thousands of proteins from cells, tissues, and biofluids, including members of the PI3K/AKT/mTOR pathway and the PD-L1 signaling axis. He furthermore developed MS assays to determine mutation rates of known cancer drivers in tumor tissues on the protein level, and to determine the concentration of immune-therapeutics in patient blood, for therapeutic drug monitoring. The targeted MS assays developed in the Borchers lab are being used both for fundamental research and to complement existing genomic assays in precision oncology. Dr. Borchers has over 325 publications in mass-spectrometry based proteomics, with an H-index of 77 and more than 25,000 citations.

(Structural Proteomics, Clinical MS, Metabolomics). He received his MD from the Second Moscow Medical Institute, Moscow, Russia, and his Ph.D. in bioorganic chemistry from the Institute of Bioorganic Chemistry of the National Academy of Sciences of Belarus, Minsk, Belarus. He conducted postdoctoral studies at the National Institute of Environmental Health Sciences, National Institutes of Health in Research Triangle Park, NC and at the University of North Carolina at Chapel Hill, NC. He was a Research Assistant Professor at the University of Victoria-Genome BC Proteomics Centre, University of Victoria, British Columbia, Canada. His research is focused on the development and application of the structural proteomics approaches for studying protein structure in pathologies such as neurodegenerative protein misfolding diseases. Since recently he is involved in the development and application of the analytical MS methods in clinics and the development of metabolomics analyses.

Yassene is an assistant professor at the Center for Proteomics and Metabolomics at Leiden University Medical Center, the Netherlands. He is also head of bioinformatics at the Proteomics Centre of the University of Victoria, Canada, where he is also an adjunct assistant professor at the Division of Medical Science. He received his PhD from the University of Göttingen, Germany. A large portion of Yassene’s recent activities focuses on quantitative targeted proteomics and its clinical applications. He has co-authored multiple works on the methods and applications of precise quantitation of blood plasma proteins using mass spectrometry and heavy labeled surrogate peptide internal standards. He is also active in perusing new approaches for mass spectrometry data analysis, -omics integration, as well as automation of information extraction from such data.

Adriano is a Senior Administrative Coordinator at the SCPC. He was an Associate Professor at the Federal University of Minas Gerais (UFMG), Brazil, for 18 years before moving to Canada in 2022 and joining the SCPC in November/22. Graduated in Biological Sciences (Bachelor in Zoology) and Ph.D. in Biological Sciences (Physiology and Pharmacology) from UFMG, Adriano was also a postdoctoral fellow at the Université de la Méditerranée (Marseille, France; 1999/2001) and visiting professor at the University of Alberta (2013-2014). He has a strong background in leading structurally- or functionally-driven drug discovery research projects combining biochemistry, molecular pharmacology, cell biology, and proteomic approaches. Adriano is responsible for overseeing and managing the operations, budgets and activities of the proteomics research and its team at the SCPC.

Dr. Richard is a Research Associate at the SCPC and project manager of a Genome Canada-funded GAPP project. He has joined the SCPC in 2015. He is developing methods for quantitative proteomics (discovery and validation) as well metabolomics (targeted metabolite quantitation). He is involved in multiple collaborative projects within the Lady Davis Institute for Medical Research and McGill University. Vincent finished his PhD in 2014 in the laboratory of Dr. Vladimir Titorenko where he used LC/MS based proteomics, lipidomics, and metabolomics to elucidate the molecular mechanisms regulating aging and lifespan of model organisms.

Deema is an experienced scientist working at the SCPC. She obtained her bachelor’s degree in Pharmacy from the Jordan University of Science and Technology. She later received a Master of Medical Science in Clinical Biochemistry from the University of Calgary. Before joining the SCPC, Deema held positions as a Laboratory Scientist at Jordan University of Science and Technology and Alberta Precision Laboratories, where her research focused on the development of mass spectrometry-based tools for early and accurate diagnosis of metabolic disorders such as Lysosomal Storage Disorders and Congenital Adrenal Hyperplasia using dried blood spots. Since joining the SCPC in 2021, Deema has contributed to the institute’s research, method development and validation and collaborative projects in clinical mass spectrometry, metabolomics, lipidomics, and proteomics.

Dennis is a research technician at the SCPC. Dennis graduated with a bachelor’s degree in Biological Sciences at the University of British Columbia. He worked as a laboratory assistant at the University of British Columbia under Dr. Shyh-Dar Li in the pharmaceutical sciences department, assisting with nanoparticle research and drug delivery projects. Dennis joined the SCPC in 2022, to aid in analytical mass spectrometry, metabolomics, and proteomics research.

Ute joined the Segal Cancer Proteomics Centre in 2019. She obtained her bachelor’s degree in Chemistry from Natural Science-Technical Academy, Isny, Germany.


Georgia Mitsa is a PhD candidate in the Department of Experimental Medicine at McGill University in Montreal, Canada. Her research combines proteomics, metabolomics and genomics to gain mechanistic insights into hard-to-treat malignancies. Georgia aims to advance the translation of multi-omics into the clinic to generate a complete phenotypic picture of oncogenic molecular changes. The ultimate goal is to enable quick and reliable clinical decision-making using multi-omics analysis of clinical biopsies. To this end, Georgia has developed several quantitative mass spectrometry-based methods for multi-omics molecular characterization of biobanked clinical samples. She is currently spearheading a large-scale study to investigate the molecular changes in metastatic colorectal cancers that are refractory to standard-of-care treatment. She further initiated and conducts a large-scale study targeting proteomic changes in pre-invasive breast ductal carcinoma; this project applies her recently-published method for mass spectrometry-based quantitative proteomics from core-needle biopsies embedded in formalin and paraffin.

Connie is a PhD candidate who joined the SCPC in 2017. She is supervised by Dr. Gerald Batist. Her project involves validating and applying quantitative proteomics technologies in cancer tissue samples to better predict patients’ individual responses to targeted treatments.

Vincent is a graduate student who has joined the SCPC in 2019. He is co-supervised by Dr. Alan Spatz, and he is working on an immuno-MS assay to quantify the PD-1/PD-L1 axis in order to better predict the response of individual tumours to immune checkpoint inhibitors. He completed his bachelor’s degree in Pharmacology (co-operative program) and Master’s degree at Université de Sherbrooke before joining the SCPC. Throughout his studies, he focused on understanding variability in treatment effectiveness, particularly for cancer. This led him to explore mass spectrometry-based proteomics to gain insight into tumour biology.

Neda is a graduate student who has joined the SCPC in 2022. She is co-supervised by Dr. Allen Spatz, and she is working on immuno-Ms assays to study PD-L1 signalling.

Forough is a graduate student who has joined the SCPC in 2022. She graduated with a MD degree from Beheshti University of Medical Science (SBMU) and she is working on structural proteomics.

Costa is graduate student who has joined the SCPC in 2022 and he is working on Bioinformatics approaches on cancer research.

Fatemeh is a graduate student who has joined the SCPC in 2022. She is co-supervised by Dr. Michael Pollak, and she is working on Characterization of Signaling Pathway Activation in Breast Cancer using Comprehensive Targeted Phosphoproteomics.

Negarsadat Mostolizadeh is a MSc. student at McGill University and has become a member of the SCPC in 2022. She is co-supervised by Dr. Mark Basik. Her current research project involves the use of the MRM assay to quantify Her2 expression in Her2-low tumors. The purpose of this work is to distinguish between HER2-low and HER2-negative cases and to provide improved information for therapeutic decision-making for cancer patients.

Neginsadat Mostolizadeh is a MSc. student at McGill University and a member of the SCPC. She joined the team in 2022 and is co-supervised by Dr. Gerald Batist. Her Master’s thesis project focuses on testing and optimizing the performance of a proteomic profile that is associated with the response of Capivasertib in cancer cell lines and patient tumor samples. Her ultimate goal is to make proteomic profile data useful in clinical settings.
Qasrawi DO, Al-Ghabkari A, Khan RM, Petrotchenko EV, Montero-Odasso M, Borchers CH.
A Simplified Proteomics LC-MRM-MS Assay for Determination of apoE Genotypes in Plasma Samples.
J Proteome Res. 2024 Feb 27. DOI: 10.1021/acs.jproteome.3c00557. Online ahead of print.
PMID: 38412507.
Luo V, Shen C, Worme S, Bhagrath A, Simo-Cheyou E, Findlay S, Hebert S, Poon W, Aryanpour Z, Zhang T, Zahedi R, Boulais J, Buchwald Z, Borchers CH, Cote J, Kleinman C, Mandl J, Orthwein A.
The Deubiquitylase Otub1 Regulates the Chemotactic Response of Splenic B Cells by Modulating the Stability of the γ-Subunit Gng2.
Molecular and Cellular Biology 2024 Jan 29; 44:1, 1-16, DOI: 10.1080/10985549.2023.2290434.
Mohammed Y, Tran K, Carlsten C, Ryerson C, Wong A, Lee T, Cheng MP, Vinh DC, Lee TC, Winston BW, Sweet D, Boyd JH, Walley KR, Haljan G, McGeer A, Lamontagne F, Fowler R, Maslove D, Singer J, Patrick DM, Marshall JC, Murthy S, Jain F, Borchers CH, Goodlett DR, Levin A, Russell JA; ARBs CORONA I Consortium.
Proteomic Evolution from Acute to Post-COVID-19 Conditions.
J Proteome Res. 2024 Jan 5;23(1):52-70. DOI: 10.1021/acs.jproteome.3c00324.
PMID: 38048423 Free PMC article.
Sobsey CA, Mady N, Richard VR, LeBlanc A, Zakharov T, Borchers CH, Jagoe RT.
Measurement of CYP1A2 and CYP3A4 activity by a simplified Geneva cocktail approach in a cohort of free-living individuals: a pilot study.
Front Pharmacol. 2024 Feb 2;15:1232595. DOI: 10.3389/fphar.2024.1232595. eCollection 2024.
PMID: 38370474 Free PMC article.
Michaud SA, Pětrošová H, Sinclair NJ, Kinnear AL, Jackson AM, McGuire JC, Hardie DB, Bhowmick P, Ganguly M, Flenniken AM, Nutter LMJ, McKerlie C, Smith D, Mohammed Y, Schibli D, Sickmann A, Borchers CH.
Multiple reaction monitoring assays for large-scale quantitation of proteins from 20 mouse organs and tissues.
Commun Biol. 2024 Jan 2;7(1):6. DOI: 10.1038/s42003-023-05687-0.
PMID: 38168632.
Reboucas P, Fillebeen C, Botta A, Cleverdon R, Steele AP, Richard VR, Zahede RP, Borchers CH, Burelle Y, Hawkes TJ, Pantopoulos K, Sweeney G.
Discovery-Based Proteomics Identify Skeletal Muscle Mitochondrial Alterations as an Early Metabolic Defect in a Mouse Model of β-Thalassemia
Int. J. Mol. Sci. 2023 Feb 23, 24(5), 4402; https://doi.org/10.3390/ijms24054402.
Mohammed Y, Goodlett D, Borchers CH.
Bioinformatics Tools and Knowledgebases to Assist Generating Targeted Assays for Plasma Proteomics
Methods Mol Biol. 2023 Feb 12; 2628: 557-577. doi: 10.1007/978-1-0716-2978-9_32.
PMID: 36781806
Mohammed Y, Goodlett D, Borchers CH.
Absolute Quantitative Targeted Proteomics Assays for Plasma Proteins
Methods Mol Biol. 2023 Feb 12; 2628: 439-473. doi: 10.1007/978-1-0716-2978-9_27.
PMID: 36781801
Prabhu SA, Moussa O, Gonçalves C, LaPierre JH, Chou H, Huang F, Richard VR, Ferruzo PYM, Guettler EM, Soria-Bretones I, Kirby L, Gagnon N, Su J, Silvester J, Krisna SS, Rose AAN, Sheppard KE, Cescon DW, Mallette FA, Zahedi RP, Borchers CH, Del Rincon SV, Miller WH.
Inhibition of the MNK1/2-eIF4E Axis Augments Palbociclib-Mediated Antitumor Activity in Melanoma and Breast Cancer
Mol Cancer Ther, 2023 Feb 1;22(2):192-204. doi: 10.1158/1535-7163.MCT-22-0092.
PMID: 36722142
Preston SEJ, Richard VR, Del Rincón SV, Borchers CH, Zahedi RP.
Proteomic Assessment of the Murine Mammary Gland Extracellular Matrix.
Methods Mol Biol, 2023;2614:261-271. doi: 10.1007/978-1-0716-2914-7_16.
PMID: 36587130
Parra ALC, Freitas CDT, Souza PFN, von Aderkas P, Borchers CH, Beattie GA, Silva FDA, Thornburg RW.
Ornamental tobacco floral nectar is a rich source of antimicrobial peptides.
Plant Sci, 2022 Nov;324:111427. doi: 10.1016/j.plantsci.2022.111427. Epub 2022 Aug 23.
PMID: 36007629
Richard VR, Gaither C, Popp R, Chaplygina D, Brzhozovskiy A, Kononikhin A, Mohammed Y, Zahedi RP, Nikolaev EN, Borchers CH.
Early Prediction of COVID-19 Patient Survival by Targeted Plasma Multi-Omics and Machine Learning.
Mol Cell Proteomics, 2022 Oct;21(10):100277. doi: 10.1016/j.mcpro.2022.100277. Epub 2022 Aug 3.
PMID: 35931319
Smith-Byrne K, Cerani A, Guida F, Zhou S, Agudo A, Aleksandrova K, Barricarte A, Rodríguez-Barranco M, Borchers CH, Gram IT, Han J, Amos CI, Hung RJ, Grankvist K, Nøst TH, Imaz L, Chirlaque-López MD, Johansson M, Kaaks R, Kühn T, Martin RM, McKay JD, Pala V, Robbins HA, Sandanger TM, Schibli D, Schulze MB, Travis RC, Vineis P, Weiderpass E, Brennan P, Johansson M, Richards JB.
Circulating Isovalerylcarnitine and Lung Cancer Risk: Evidence from Mendelian Randomization and Pre-Diagnostic Blood Measurements.
Cancer Epidemiol Biomarkers Prev, 2022 Jul 15:EPI-21-1033. doi: 10.1158/1055-9965.EPI-21-1033. Online ahead of print.
PMID: 35839461
Preston SEJ, Bartish M, Richard VR, Aghigh A, Gonçalves C, Smith-Voudouris J, Huang F, Thebault P, Cleret-Buhot A, Lapointe R, Légaré F, Postovit LM, Zahedi RP, Borchers CH, Miller WH Jr, Del Rincón SV.
Phosphorylation of eIF4E in the stroma drives the production and spatial organisation of collagen type I in the mammary gland.
Matrix Biol, 2022 Jul 13:S0945-053X(22)00092-0. doi: 10.1016/j.matbio.2022.07.003. Online ahead of print.
PMID: 35842012
Pastushkova LK, Goncharov IN, Koloteva MI, Goncharova AG, Kashirina DN, Nosovsky AM, Glebova TM, Kononikhin AS, Borchers CH, Nikolaev EN, Larina IM.
Characteristics of blood plasma proteome changes associated with the hemorrhagic purpura of cosmonauts on the first day after long-term space missions.
Life Sci Space Res (Amst), 2022 May;33:7-12. doi: 10.1016/j.lssr.2022.01.001. Epub 2022 Jan 14.
PMID: 35491032
Mitsa G, Guo Q, Goncalves C, Preston SEJ, Lacasse V, Aguilar-Mahecha A, Benlimame N, Basik M, Spatz A, Batist G, Miller WH Jr, Del Rincon SV, Zahedi RP, Borchers CH.
A Non-Hazardous Deparaffinization Protocol Enables Quantitative Proteomics of Core Needle Biopsy-Sized Formalin-Fixed and Paraffin-Embedded (FFPE) Tissue Specimens.
Int J Mol Sci, 2022 Apr 18;23(8):4443. doi: 10.3390/ijms23084443.
PMID: 35457260
Laselva O, Petrotchenko EV, Hamilton CM, Qureshi Z, Borchers CH, Young RN, Bear CE.
A protocol for identifying the binding sites of small molecules on the cystic fibrosis transmembrane conductance regulator (CFTR) protein.
STAR Protoc, 2022 Apr 7;3(2):101258. doi: 10.1016/j.xpro.2022.101258. eCollection 2022 Jun 17.
PMID: 35434660
Mohammed Y, Touw CE, Nemeth B, van Adrichem RA, Borchers CH, Rosendaal FR, van Vlijmen BJ, Cannegieter SC.
Targeted proteomics for evaluating risk of venous thrombosis following traumatic lower-leg injury or knee arthroscopy.
J Thromb Haemost, 2022 Mar;20(3):684-699. doi: 10.1111/jth.15623. Epub 2022 Jan 6.
PMID: 34919779
Gaither C, Popp R, Zahedi RP, Borchers CH.
Multiple reaction monitoring-mass spectrometry enables robust quantitation of plasma proteins regardless of whole blood processing delays that may occur in the clinic.
Mol Cell Proteomics, 2022 May;21(5):100212. doi: 10.1016/j.mcpro.2022.100212. Epub 2022 Feb 17.
PMID: 35182769
Jennings MJ, Kagiava A, Vendredy L, Spaulding EL, Stavrou M, Hathazi D, Grüneboom A, De Winter V, Gess B, Schara U, Pogoryelova O, Lochmüller H, Borchers CH, Roos A, Burgess RW, Timmerman V, Kleopa KA, Horvath R.
NCAM1 and GDF15 are biomarkers of Charcot-Marie-Tooth disease in patients and mice.
Brain, 2022 Nov 21;145(11):3999-4015. doi: 10.1093/brain/awac055.
PMID: 35148379
Brzhozovskiy A, Kononikhin A, Bugrova AE, Kovalev GI, Schmit PO, Kruppa G, Nikolaev EN, Borchers CH.
The Parallel Reaction Monitoring-Parallel Accumulation-Serial Fragmentation (prm-PASEF) Approach for Multiplexed Absolute Quantitation of Proteins in Human Plasma.
Anal Chem, 2022 Jan 18. doi: 10.1021/acs.analchem.1c03782. Online ahead of print.
PMID: 35040635
Petrotchenko EV, Borchers CH.
Protein Chemistry Combined with Mass Spectrometry for Protein Structure Determination.
Chem Rev, 2022 Apr 27;122(8):7488-7499. doi: 10.1021/acs.chemrev.1c00302. Epub 2021 Dec 30.
PMID: 34968047
Gaspar VP, Ibrahim S, Zahedi RP, Borchers CH.
Utility, Promise, and Limitations of LC-MS based Therapeutic Drug Monitoring in Precision Medicine.
Journal of Mass Spectrometry, 2021 Nov;56(11):e4788. doi: 10.1002/jms.4788. Epub 2021 Nov 4.
PMID: 34738286 Review
Froehlich BC, Popp R, Sobsey CA, Ibrahim S, LeBlanc A, Mohammed Y, Buchanan M, Aguilar-Mahecha A, Pötz O, Chen MX, Spatz A, Basik M, Batist G, Zahedi RP, Borchers CH.
A multiplexed, automated immuno-matrix assisted laser desorption/ionization mass spectrometry assay for simultaneous and precise quantitation of PTEN and p110α in cell lines and tumor tissues.
Analyst, 2021 Oct 25, 146, 6566-6575 doi: 10.1039/d1an00165e. Epub 2021 Sep 29.
PMID: 34585690
Serpa JJ, Popov KI, Petrotchenko EV, Dokholyan NV, Borchers CH.
Structure of prion β-oligomers as determined by short-distance crosslinking constraint-guided discrete molecular dynamics simulations.
Proteomics, 2021 Sep 5:e2000298. doi: 10.1002/pmic.202000298. Online ahead of print.
PMID: 34482645
Ibrahim S, Lan C, Chabot C, Mitsa G, Buchanan M, Aguilar-Mahecha A, Elchebly M, Poetz O, Spatz A, Basik M, Batist G, Zahedi RP, Borchers CH.
Precise Quantitation of PTEN by Immuno-MRM: A Tool to Resolve the Breast Cancer Biomarker Controversy.
Anal Chem, 2021 Aug 10;93(31):10816-10824. doi: 10.1021/acs.analchem.1c00975. Epub 2021 Jul 29.
PMID: 34324311
Gaither C, Popp R, Borchers SP, Skarphedinsson K, Eiriksson FF, Thorsteinsdóttir M, Mohammed Y, Borchers CH.
Performance Assessment of a 125 Human Plasma Peptide Mixture Stored at Room Temperature for Multiple Reaction Monitoring-Mass Spectrometry.
J Proteome Res, 2021 Sep 3;20(9):4292-4302. doi: 10.1021/acs.jproteome.1c00249. Epub 2021 Jul 16.
PMID: 34270269
Skinnider MA, Rogalski J, Tigchelaar S, Manouchehri N, Prudova A, Jackson AM, Nielsen K, Jeong J, Chaudhary S, Shortt K, Gallagher-Kurtzke Y, So K, Fong A, Gupta R, Okon EB, Rizzuto MA, Dong K, Streijger F, Belanger L, Ritchie L, Tsang A, Christie S, Mac-Thiong JM, Bailey C, Ailon T, Charest-Morin R, Dea N, Wilson JR, Dhall S, Paquette S, Street J, Fisher CG, Dvorak MF, Shannon C, Borchers C, Balshaw R, Foster LJ, Kwon BK.
Proteomics portraits reveal evolutionarily conserved and divergent responses to spinal cord injury.
Mol Cell Proteomics, 2021 Jun 12:100096. doi: 10.1016/j.mcpro.2021.100096. Online ahead of print.
PMID: 34129941
Tran JN, Günther OP, Sherwood KR, Fenninger F, Allan LL, Lan J, Sapir-Pichhadze R, Duquesnoy R, Claas F, Marsh SGE, McMaster WR, Keown PA; Genome Canada Transplant Consortium.
High-throughput sequencing defines donor and recipient HLA B-cell epitope frequencies for prospective matching in transplantation.
Commun Biol, 2021 May 14;4(1):583. doi: 10.1038/s42003-021-01989-3.
PMID: 33990681
Laselva O, Qureshi Z, Zeng ZW, Petrotchenko EV, Ramjeesingh M, Hamilton CM, Huan LJ, Borchers CH, Pomès R, Young R, Bear CE.
Identification of binding sites for ivacaftor on the cystic fibrosis transmembrane conductance regulator.
iScience, 2021 May 15;24(6):102542. doi: 10.1016/j.isci.2021.102542. eCollection 2021 Jun 25.
PMID: 34142049
Mohammed Y, Michaud SA, Pětrošová H, Yang J, Ganguly M, Schibli D, Flenniken AM, Nutter LMJ, Adissu HA, Lloyd KCK, McKerlie C, Borchers CH.
Proteotyping of knockout mouse strains reveals sex- and strain-specific signatures in blood plasma.
NPJ Syst Biol Appl, 2021 May 28;7(1):25. doi: 10.1038/s41540-021-00184-8.
PMID: 34050187
Li FKK, Gale RT, Petrotchenko EV, Borchers CH, Brown ED, Strynadka NCJ.
Crystallographic analysis of TarI and TarJ, a cytidylyltransferase and reductase pair for CDP-ribitol synthesis in Staphylococcus aureus wall teichoic acid biogenesis.
J Struct Biol, 2021 Jun;213(2):107733. doi: 10.1016/j.jsb.2021.107733. Epub 2021 Apr 2.
PMID: 33819634
Bhowmick P, Roome S, Borchers CH, Goodlett DR, Mohammed Y.
An Update on MRMAssayDB: A Comprehensive Resource for Targeted Proteomics Assays in the Community.
Proteome Res, 2021 Apr 2;20(4):2105-2115. doi: 10.1021/acs.jproteome.0c00961. Epub 2021 Mar 8.
PMID: 33683131
Mohammed Y, Bhowmick P, Michaud SA, Sickmann A, Borchers CH.
Mouse Quantitative Proteomics Knowledgebase: reference protein concentration ranges in 20 mouse tissues using 5000 quantitative proteomics assays.
Bioinformatics, 2021 Jan 23:btab018. doi: 10.1093/bioinformatics/btab018. Online ahead of print.
PMID: 33483739
Shahid I, Han J, Hardie D, Baig DN, Malik KA, Borchers CH, Mehnaz S.
Profiling of antimicrobial metabolites of plant growth promoting Pseudomonas spp. isolated from different plant hosts.
3 Biotech, 2021 Feb;11(2):48. doi: 10.1007/s13205-020-02585-8. Epub 2021 Jan 11.
PMID: 33489669
Michaud SA, Pětrošová H, Jackson AM, McGuire JC, Sinclair NJ, Ganguly M, Flenniken AM, Nutter LMJ, McKerlie C, Schibli D, Smith D, Borchers CH.
Process and Workflow for Preparation of Disparate Mouse Tissues for Proteomic Analysis.
J Proteome Res, 2021 Jan 1;20(1):305-316. doi: 10.1021/acs.jproteome.0c00399. Epub 2020 Nov 5.
PMID: 33151080
Gaspar VP, Ibrahim S, Sobsey CA, Richard VR, Spatz A, Zahedi RP, Borchers CH.
Direct and Precise Measurement of Bevacizumab Levels in Human Plasma Based on Controlled Methionine Oxidation and Multiple Reaction Monitoring.
ACS Pharmacol Transl Sci, 2020 Nov 13;3(6):1304-1309. doi: 10.1021/acsptsci.0c00134. eCollection 2020 Dec 11.
PMID: 33344903
De Giorgi M, Jarrett KE, Burton JC, Doerfler AM, Hurley A, Li A, Hsu RH, Furgurson M, Patel KR, Han J, Borchers CH, Lagor WR.
Depletion of essential isoprenoids and ER stress induction following acute liver-specific deletion of HMG-CoA reductase.
J Lipid Res, 2020 Dec;61(12):1675-1686. doi: 10.1194/jlr.RA120001006. Epub 2020 Oct 27.
PMID: 33109681
Leitner A, Bonvin AMJJ, Borchers CH, Chalkley RJ, Chamot-Rooke J, Combe CW, Cox J, Dong MQ, Fischer L, Götze M, Gozzo FC, Heck AJR, Hoopmann MR, Huang L, Ishihama Y, Jones AR, Kalisman N, Kohlbacher O, Mechtler K, Moritz RL, Netz E, Novak P, Petrotchenko E, Sali A, Scheltema RA, Schmidt C, Schriemer D, Sinz A, Sobott F, Stengel F, Thalassinos K, Urlaub H, Viner R, Vizcaíno JA, Wilkins MR, Rappsilber J.
Toward Increased Reliability, Transparency, and Accessibility in Cross-linking Mass Spectrometry.
Structure, 2020 Nov 3;28(11):1259-1268. doi: 10.1016/j.str.2020.09.011. Epub 2020 Oct 15.
PMID: 33065067
Richard VR, Zahedi RP, Eintracht S, Borchers CH.
An LC-MRM assay for the quantification of metanephrines from dried blood spots for the diagnosis of pheochromocytomas and paragangliomas.
Anal Chim Acta, 2020 Sep 1;1128:140-148. doi: 10.1016/j.aca.2020.06.020. Epub 2020 Jul 9.
PMID: 32825898
Ibrahim S, Froehlich B, Aguilar-Mahecha A, Aloyz R, Poetz O, Basik M, Batist G, Zahedi RP, Borchers CH.
Using two peptide isotopologues as internal standards for the streamlined quantification of low-abundance proteins by immuno-MRM and immuno-MALDI.
Anal Chem, 2020 Aug 12. doi: 10.1021/acs.analchem.0c02157. Online ahead of print.
PMID: 32786432
Nikolaev EN, Indeykina MI, Brzhozovskiy AG, Bugrova AE, Kononikhin AS, Starodubtseva NL, Petrotchenko EV, Kovalev GI, Borchers CH, Sukhikh GT.
Mass Spectrometric detection of SARS-CoV-2 virus in scrapings of the epithelium of the nasopharynx of infected patients via Nucleocapsid N protein.
J Proteome Res, 2020 Aug 19. doi: 10.1021/acs.jproteome.0c00412. Online ahead of print.
PMID: 32786682
Froehlich BC, Popp R, Sobsey CA, Ibrahim S, LeBlanc AM, Mohammed Y, Aguilar-Mahecha A, Poetz O, Chen MX, Spatz A, Basik M, Batist G, Zahedi RP, Borchers CH.
Systematic Optimization of the iMALDI Workflow for the Robust and Straightforward Quantification of Signaling Proteins in Cancer Cells.
Proteomics Clin Appl, 2020 Jul 8:e2000034. doi: 10.1002/prca.202000034. Online ahead of print.
PMID: 32643306
Kashirina DN, Brzhozovskiy AG, Pastushkova LKh, Kononikhin AS, Borchers CH, Nikolaev EN, Larina IM.
Semiquantitative Proteomic Research of Protein Plasma Profile of Volunteers in 21-Day Head-Down Bed Rest.
Front Physiol, 2020 Jul 24;11:678. doi: 10.3389/fphys.2020.00678. eCollection 2020.
PMID: 32848806
Liu T, Wang RX, Han J, Qiu YL, Borchers CH, Ling V, Wang JS.
Changes in plasma bile acid profiles after partial internal biliary diversion in PFIC2 patients.
Ann Transl Med, 2020 Mar;8(5):185. doi: 10.21037/atm.2020.01.103.
PMID: 32309332
Sobsey CA, Ibrahim S, Richard VR, Gaspar V, Mitsa G, Lacasse V, Zahedi RP, Batist G, Borchers CH.
Targeted and Untargeted Proteomics Approaches in Biomarker Development.
Proteomics. 2020 May;20(9):e1900029. doi: 10.1002/pmic.201900029. Epub 2020 Jan 30.
PMID: 31729135
Review
Gaither C, Popp R, Mohammed Y, Borchers CH.
Determination of the concentration range for 267 proteins from 21 lots of commercial human plasma using highly multiplexed multiple reaction monitoring mass spectrometry.
Analyst, 2020 May 18;145(10):3634-3644. doi: 10.1039/c9an01893j.
PMID: 32255452
Tilburg J, Michaud SA, Maracle CX, Versteeg HH, Borchers CH, van Vlijmen BJM, Mohammed Y.
Plasma Protein Signatures of a Murine Venous Thrombosis Model and Slc44a2 Knockout Mice Using Quantitative-Targeted Proteomics.
Thromb Haemost, 2020 Mar;120(3):423-436. doi: 10.1055/s-0040-1702229. Epub 2020 Mar 5.
PMID: 32135565
Makepeace KAT, Mohammed Y, Rudashevskaya EL, Petrotchenko EV, Voegtle FN, Meisinger C, Sickmann A, Borchers CH.
Improving identification of in-organello protein-protein interactions using an affinity-enrichable, isotopically-coded, and mass spectrometry-cleavable chemical crosslinker.
Mol Cell Proteomics, 2020 Apr;19(4):624-639. doi: 10.1074/mcp.RA119.001839. Epub 2020 Feb 12.
PMID: 32051233
Bhardwaj M, Weigl K, Tikk K, Holland-Letz T, Schrotz-King P, Borchers CH, Brenner H.
Multiplex quantitation of 270 plasma protein markers to identify a signature for early detection of colorectal cancer.
Eur J Cancer, 2020 Mar;127:30-40. doi: 10.1016/j.ejca.2019.11.021. Epub 2020 Jan 21.
PMID: 31972396
Shin JJH, Liu P, Chan LJ, Ullah A, Pan J, Borchers CH, Burke JE, Stefan C, Smits GJ, Loewen CJR.
pH Biosensing by PI4P Regulates Cargo Sorting at the TGN.
Dev Cell, 2020 Feb 24;52(4):461-476.e4. doi: 10.1016/j.devcel.2019.12.010. Epub 2020 Jan 9.
PMID: 31928972
Eshghi A, Pistawka AJ, Liu J, Chen M, Sinclair NJT, Hardie DB, Elliott M, Chen L, Newman R, Mohammed Y, Borchers CH.
Concentration determination of >200 proteins in dried blood spots for biomarker discovery and validation.
Mol Cell Proteomics, 2020 Mar;19(3):540-553. doi:10.1074/mcp.TIR119.001820. Epub 2020 Jan 2.
PMID: 31896676
Silva F, Guirgis A, von Aderkas P, Borchers CH, Thornburg R.
LC-MS/MS based comparative proteomics of floral nectars reveal different mechanisms involved in floral defense of Nicotiana spp., Petunia hybrida and Datura stramonium.
J Proteomics, 2020 Feb 20;213:103618. doi: 10.1016/j.jprot.2019.103618. Epub 2019 Dec 14.
PMID: 31846763
Mohammed Y, Kootte RS, Kopatz WF, Borchers CH, Büller HR, Versteeg HH, Nieuwdorp M, van Mens TE.
The intestinal microbiome potentially affects thrombin generation in human subjects.
J Thromb Haemost, 2020 Mar;18(3):642-650. doi: 10.1111/jth.14699. Epub 2019 Dec 27.
PMID: 31808596
Sobsey CA, Ibrahim S, Richard VR, Gaspar V, Mitsa G, Lacasse V, Zahedi RP, Batist G, Borchers CH.
Targeted and Untargeted Proteomics Approaches in Biomarker Development.
Proteomics, 2020 May;20(9):e1900029. doi: 10.1002/pmic.201900029. Epub 2020 Jan 30.
PMID: 31729135
Makepeace KAT, Brodie NI, Popov KI, Gudavicius G, Nelson CJ, Petrotchenko EV, Dokholyan NV, Borchers CH.
Ligand-induced disorder-to-order transitions characterized by structural proteomics and molecular dynamics simulations.
J Proteomics, 2020 Jan 16;211:103544. doi: 10.1016/j.jprot.2019.103544. Epub 2019 Nov 1.
PMID: 31683063
Blank-Landeshammer B, Richard VR, Mitsa G, Marques M, LeBlanc A, Kollipara L, Feldmann I, Couetoux du Tertre M, Gambaro K, McNamara S, Spatz A, Zahedi RP, Sickmann A, Batist G, Borchers CH.
Proteogenomics of Colorectal Cancer Liver Metastases: Complementing Precision Oncology with Phenotypic Data.
Cancer, 2019 Dec 1;11(12):1907. doi: 10.3390/cancers11121907.
PMID: 31805664
Popov KI, Makepeace KAT, Petrotchenko EV, Dokholyan NV, Borchers CH.
Insight into the Structure of the “Unstructured” Tau Protein.
Structure, 2019 Nov 5;27(11):1710-1715.e4. doi: 10.1016/j.str.2019.09.003. Epub 2019 Oct 15.
PMID: 31628033
Martínez de Paz A, Khajavi L, Martin H, Claveria-Gimeno R, Tom Dieck S, Cheema MS, Sanchez-Mut JV, Moksa MM, Carles A, Brodie NI, Sheikh TI, Freeman ME, Petrotchenko EV, Borchers CH, Schuman EM, Zytnicki M, Velazquez-Campoy A, Abian O, Hirst M, Esteller M, Vincent JB, Malnou CE, Ausió J.
MeCP2-E1 isoform is a dynamically expressed, weakly DNA-bound protein with different protein and DNA interactions compared to MeCP2-E2.
Epigenetics Chromatin, 2019 Oct 10;12(1):63. doi: 10.1186/s13072-019-0298-1.
PMID: 31601272
Bhardwaj M, Gies A, Weigl K, Tikk K, Benner A, Schrotz-King P, Borchers CH, Brenner H.
Evaluation and Validation of Plasma Proteins Using Two Different Protein Detection Methods for Early Detection of Colorectal Cancer.
Cancers (Basel), 2019 Sep 25;11(10):1426. doi: 10.3390/cancers11101426.
PMID: 31557860
Pires ES, Hardoim CCP, Miranda KR, Secco DA, Lobo LA, de Carvalho DP, Han J, Borchers CH, Ferreira RBR, Salles JF, Domingues RMCP, Antunes LCM.
The Gut Microbiome and Metabolome of Two Riparian Communities in the Amazon.
Front Microbiol, 2019 Sep 4;10:2003. doi:10.3389/fmicb.2019.02003. eCollection 2019.
PMID: 31555238
Sirois I, Aguilar-Mahecha A, Lafleur J, Fowler E, Vu V, Scriver M, Buchanan M, Chabot C, Ramanathan A, Balachandran B, Legare S, Przybytkowski E, Lan C, Krzemien U, Cavallone L, Aleynikova O, Ferrario C, Guilbert MC, Benlimame N, Saad A, Alaoui-Jamali M, Saragovi HU, Josephy S, O’Flanagan C, Hursting SD, Richard VR, Zahedi RP, Borchers CH, Bareke E, Nabavi S, Tonellato P, Roy JA, Robidoux A, Marcus EA, Mihalcioiu C, Majewski J, Basik M.
A unique morphological phenotype in chemoresistant triple negative breast cancer reveals metabolic reprogramming and PLIN4 expression as a molecular vulnerability.
Mol Cancer Res, 2019 Dec;17(12):2492-2507. doi: 10.1158/1541-7786.MCR-19-0264. Epub 2019 Sep 19.
PMID: 31537618
Oyama Y, Bartman CM, Bonney S, Lee JS, Walker LA, Han J, Borchers CH, Buttrick PM, Aherne CM, Clendenen N, Colgan SP, Eckle T.
Intense Light-Mediated Circadian Cardioprotection via Transcriptional Reprogramming of the Endothelium.
Cell Rep, 2019 Aug 6;28(6):1471-1484.e11. doi: 10.1016/j.celrep.2019.07.020.
PMID: 31390562
Mao F, Liu T, Hou X, Zhao H, He W, Li C, Jing Z, Sui J, Wang F, Liu X, Han J, Borchers CH, Wang JS, Li W.
Increased sulfation of bile acids in mice and human subjects with sodium taurocholate cotransporting polypeptide deficiency.
J Biol Chem, 2019 Aug 2;294(31):11853-11862. doi: 10.1074/jbc.RA118.007179. Epub 2019 Jun 14.
PMID: 31201272
Masson GR, Burke JE, Ahn NG, Anand GS, Borchers C, Brier S, Bou-Assaf GM, Engen JR, Englander SW, Faber J, Garlish R, Griffin PR, Gross ML, Guttman M, Hamuro Y, Heck AJR, Houde D, Iacob RE, Jørgensen TJD, Kaltashov IA, Klinman JP, Konermann L, Man P, Mayne L, Pascal BD, Reichmann D, Skehel M, Snijder J, Strutzenberg TS, Underbakke ES, Wagner C, Wales TE, Walters BT, Weis DD, Wilson DJ, Wintrode PL, Zhang Z, Zheng J, Schriemer DC, Rand KD.
Recommendations for performing, interpreting, and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments.
Nat Methods, 2019 Jul;16(7):595-602. doi: 10.1038/s41592-019-0459-y. Epub 2019 Jun 27.
PMID: 31249422
Anwar MA, Dai DL, Wilson-McManus J, Smith D, Francis GA, Borchers CH, McManus BM, Hill JS, Cohen Freue GV.
Multiplexed LC/ESI-MRM-MS-based Assay for Identification of Coronary Artery Disease Biomarkers in Human Plasma.
Proteomics Clin Appl, 2019 Jul;13(4):e1700111. doi: 10.1002/prca.201700111. Epub 2019 Jan 28.
PMID: 30632678
Iacobucci C, Piotrowski C, Aebersold R, Amaral BC, Andrews PC, Bernfur K, Borchers CH, Brodie NI, Bruce JE, Cao Y, Chaignepain S, Chavez JD, Claverol S, Cox J, Davis TN, Degliesposti G, Dong MQ, Edinger N, Emanuelsson C, Gay M, Götze M, Gomes-Neto F, Gozzo FC, Gutierrez CB, Haupt C, Heck AJR, Herzog F, Huang L, Hoopmann MR, Kalisman N, Klykov O, Kukačka Z, Liu F, MacCoss MJ, Mechtler K, Mesika R, Moritz RL, Nagaraj N, Nesati V, Neves-Fereira AGC, Ninnis R, Novák P, O’Reilly FJ, Pelzing M, Petrotchenko EV, Piersimoni L, Plasencia M, Pukala TL, Rand KD, Rappsilber J, Reichman D, Sailer C, Sarnowski CP, Scheltema RA, Schmidt C, Schriemer DC, Shi Y, Skehel JM, Slavin M, Sobott F, Solis-Mezarino V, Stephanowitz H, Stengel F, Stieger CE, Trabjerg E, Trnka MJ, Vilaseca M, Viner R, Xiang Y, Yılmaz Ş, Zelter A, Ziemianowicz DS, Leitner A, Sinz A.
The First Community-Wide, Comparative Cross-linking Mass Spectrometry Study.
Anal Chem, 2019 Jun 4;91(11):6953-6961. doi: 10.1021/acs.analchem.9b00658. Epub 2019 May 22.
PMID: 31045356
Brodie NI, Popov KI, Petrotchenko EV, Dokholyan NV, Borchers CH.
Conformational ensemble of native α-synuclein in solution as determined by short-distance crosslinking constraint-guided discrete molecular dynamics simulations.
PLoS Comput Biol, 2019 Mar 27;15(3):e1006859. doi: 10.1371/journal.pcbi.1006859. eCollection 2019 Mar.
PMID: 30917118
Kashirina DN, Percy AJ, Pastushkova LK, Borchers CH, Kireev KS, Ivanisenko VA, Kononikhin AS, Nikolaev EN, Larina IM.
The molecular mechanisms driving physiological changes after long duration space flights revealed by quantitative analysis of human blood proteins.
BMC Med Genomics, 2019 Mar 13;12(Suppl 2):45. doi: 10.1186/s12920-019-0490-y.
PMID: 30871558
Teixeira F, Tse E, Castro H, Makepeace KAT, Meinen BA, Borchers CH, Poole LB, Bardwell JC, Tomás AM, Southworth DR, Jakob U.
Chaperone activation and client binding of a 2-cysteine peroxiredoxin.
Nat Commun, 2019 Feb 8;10(1):659. doi: 10.1038/s41467-019-08565-8.
PMID: 30737390
Shultis D, Mitra P, Huang X, Johnson J, Khattak NA, Gray F, Piper C, Czajka J, Hansen L, Wan B, Chinnaswamy K, Liu L, Wang M, Pan J, Stuckey J, Cierpicki T, Borchers CH, Wang S, Lei M, Zhang Y.
Changing the Apoptosis Pathway through Evolutionary Protein Design.
J Mol Biol, 2019 Feb 15;431(4):825-841. doi: 10.1016/j.jmb.2018.12.016. Epub 2019 Jan 6.
PMID: 30625288
Wang R, Sheps JA, Liu L, Han J, Chen PSK, Lamontagne J, Wilson PD, Welch I, Borchers CH, Ling V.
Hydrophilic bile acids prevent liver damage caused by lack of biliary phospholipid in Mdr2-/-mice.
J Lipid Res, 2019 Jan;60(1):85-97. doi: 10.1194/jlr.M088070. Epub 2018 Nov 11.
PMID: 30416103
Boczonadi V, King MS, Smith AC, Olahova M, Bansagi B, Roos A, Eyassu F, Borchers C, Ramesh V, Lochmüller H, Polvikoski T, Whittaker RG, Pyle A, Griffin H, Taylor RW, Chinnery PF, Robinson AJ, Kunji ERS, Horvath R.
Correction: Mitochondrial oxodicarboxylate carrier deficiency is associated with mitochondrial DNA depletion and spinal muscular atrophy-like disease.
Genet Med, 2019 Sep;21(9):2163-2164. doi: 10.1038/s41436-019-0506-1.
PMID: 31028354
Popp R, Li H, Borchers CH.
Immuno-MALDI (iMALDI) mass spectrometry for the analysis of proteins in signaling pathways.
Expert Rev Proteomics, 2018 Sep;15(9):701-708. doi: 10.1080/14789450.2018.1516147. Epub 2018 Sep 17.
PMID: 30169113
Han J, Higgins R, Lim MD, Atkinson K, Yang J, Lin K, Borchers CH.
Isotope-labeling derivatization with 3-nitrophenylhydrazine for LC/multiple-reaction monitoring-mass-spectrometry-based quantitation of carnitines in dried blood spots.
Anal Chim Acta, 2018 Dec 11;1037:177-187. doi: 10.1016/j.aca.2018.01.045. Epub 2018 Feb 5.
PMID: 30292292
Michaud SA, Sinclair NJ, Pětrošová H, Palmer AL, Pistawka AJ, Zhang S, Hardie DB, Mohammed Y, Eshghi A, Richard VR, Sickmann A, Borchers CH.
Molecular phenotyping of laboratory mouse strains using 500 multiple reaction monitoring mass spectrometry plasma assays.
Common Biol, 2018 Jun 27;1:78. doi: 10.1038/s42003-018-0087-6. eCollection 2018.
PMID: 30271959
Zahedi RP, Parker CE, Borchers CH.
Immuno-MALDI-TOF-MS in the Clinic.
Clin Chem, 2018 Sep;64(9):1271-1272. doi: 10.1373/clinchem.2018.292136. Epub 2018 Jul 17.
PMID: 30018057
Penn AM, Bibok MB, Saly VK, Coutts SB, Lesperance ML, Balshaw RF, Votova K, Croteau NS, Trivedi A, Jackson AM, Hegedus J, Klourfeld E, Yu AYX, Zerna C, Modi J, Barber PA, Hoag G, Borchers CH; SpecTRA study group.
Validation of a Proteomic Biomarker Panel to Diagnose Minor-Stroke and Transient Ischaemic Attack: Phase 2 of SpecTRA, a Large-Scale Translational Study.
Biomarkers, 2018 Dec;23(8):793-803. doi: 10.1080/1354750X.2018.1499130. Epub 2018 Aug 23.
PMID: 30010432
Wölter M, Okai CA, Smith D, Ruß M, Rath W, Pecks U, Borchers CH, Glocker MO.
Maternal Apolipoprotein B100 Serum Levels are Diminished in Pregnancies with Intrauterine Growth Restriction and Differentiate from Controls.
Proteomics Clin Appl, 2018 Nov;12(6):e1800017. doi: 10.1002/prca.201800017. Epub 2018 Jul 23.
PMID: 29956482
Mahdavi S, Jenkins DJA, Borchers CH, El-Sohemy A.
Genetic Variation in 9p21 and the Plasma Proteome.
J Proteome Res, 2018 Aug 3;17(8):2649-2656. doi: 10.1021/acs.jproteome.8b00117. Epub 2018 Jul 19.
PMID: 29905076
Mnatsakanyan R, Shema G, Basik M, Batist G, Borchers CH, Sickmann A, Zahedi RP.
Detecting post-translational modification signatures as potential biomarkers in clinical mass spectrometry.
Expert Rev Proteomics, 2018 Jun;15(6):515-535. doi: 10.1080/14789450.2018.1483340.
PMID: 29893147
Bhowmick P, Mohammed Y, Borchers CH.
MRMAssayDB: an integrated resource for validated targeted proteomics assays.
Bioinformatics, 2018 Oct 15;34(20):3566-3571. doi: 10.1093/bioinformatics/bty385.
PMID: 29762640
Popp R, Basik M, Spatz A, Batist G, Zahedi RP, Borchers CH.
How iMALDI can improve clinical diagnostics.
Analyst, 2018 May 15;143(10):2197-2203. doi: 10.1039/c8an00094h.
PMID: 29713694
Parker CE, Borchers CH.
The Special Issue: Clinical Proteomics for Precision Medicine.
Proteomics Clin Appl, 2018 Mar;12(2). doi: 10.1002/prca.201600144.
PMID: 29521035
Luehr TC, Koide EM, Wang X, Han J, Borchers CH, Helbing CC.
Metabolomic insights into the effects of thyroid hormone on Rana [Lithobates] catesbeiana metamorphosis using whole-body Matrix Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging (MALDI-MSI).
Gen Comp Endocrinol, 2018 Sep 1;265:237-245. doi: 10.1016/j.ygcen.2018.02.012. Epub 2018 Feb 19.
PMID: 29470956
Liu T, Wang RX, Han J, Hao CZ, Qiu YL, Yan YY, Li LT, Wang NL, Gong JY, Lu Y, Zhang MH, Xie XB, Yang JC, You YJ, Li JQ, Knisely AS, Borchers CH, Ling V, Wang JS.
Comprehensive bile acid profiling in hereditary intrahepatic cholestasis: Genetic and clinical correlations.
Liver Int, 2018 Sep;38(9):1676-1685. doi: 10.1111/liv.13714. Epub 2018 Mar 12.
PMID: 29412511
Penn AM, Bibok MB, Saly VK, Coutts SB, Lesperance ML, Balshaw RF, Votova K, Croteau NS, Trivedi A, Jackson AM, Hegedus J, Klourfeld E, Yu AYX, Zerna C, Borchers CH.
Verification of a Proteomic Biomarker Panel to Diagnose Minor Stroke and Transient Ischaemic Attack: Phase 1 of SpecTRA, a Large-Scale Translational Study.
Biomarkers, 2018 May-Jun;23(4):392-405. doi: 10.1080/1354750X.2018.1434681. Epub 2018 Feb 12.
PMID: 29385837
Penn AM, Saly V, Trivedi A, Lesperance ML, Votova K, Jackson AM, Croteau NS, Balshaw RF, Bibok MB, Smith DS, Lam KK, Morrison J, Lu L, Coutts SB, Borchers CH.
Differential Proteomics for Distinguishing Ischemic Stroke from Controls: a Pilot Study of the SpecTRA Project.
Transl Stroke Res, 2018 Dec;9(6):590-599. doi: 10.1007/s12975-018-0609-z. Epub 2018 Jan 24.
PMID: 29368175
Dilworth D, Gudavicius G, Xu X, Boyce AKJ, O’Sullivan C, Serpa JJ, Bilenky M, Petrochenko EV, Borchers CH, Hirst M, Swayne LA, Howard P, Nelson CJ.
The prolyl isomerase FKBP25 regulates microtubule polymerization impacting cell cycle progression and genomic stability.
Nucleic Acids Res, 2018 Mar 16;46(5):2459-2478. doi: 10.1093/nar/gky008.
PMID: 29361176
Perzanowska A, Fatalska A, Wojtas G, Lewandowicz A, Michalak A, Krasowski G, Borchers CH, Dadlez M, Domanski D.
An MRM-Based Cytokeratin Marker Assay as a Tool for Cancer Studies: Application to Lung-Cancer Pleural Effusions.
Proteomics Clin Appl, 2018 Mar;12(2). doi: 10.1002/prca.201700084. Epub 2018 Feb 7.
PMID: 29352525
Brodie NI, Huguet R, Zhang T, Viner R, Zabrouskov V, Pan J, Petrotchenko EV, Borchers CH.
Top-down hydrogen-deuterium exchange analysis of protein structures using ultraviolet photodissociation (UVPD).
Anal Chem, 2018 Mar 6;90(5):3079-3082. doi: 10.1021/acs.analchem.7b03655. Epub 2018 Feb 1.
PMID: 29336549
Jardim A, Hardie DB, Boitz J, Borchers CH.
Proteomic Profiling of Leishmania donovani Promastigote Subcellular Organelles.
J Proteome Res, 2018 Mar 2;17(3):1194-1215. doi: 10.1021/acs.jproteome.7b00817. Epub 2018 Feb 5.
PMID: 29332401
Han J, Higgins R, Lim MD, Lin K, Yang J, Borchers CH.
Short-Term Stabilities of 21 Amino Acids in Dried Blood Spot.
Clin Chem, 2018 Feb;64(2):400-402. doi: 10.1373/clinchem.2017.278457. Epub 2017 Nov 2.
PMID: 29097508
Bowden JA, Heckert A, Ulmer CZ, Jones CM, Koelmel JP, Abdullah L, Ahonen L, Alnouti Y, Armando A, Asara JM, Bamba T, Barr JR, Bergquist J, Borchers CH, Brandsma J, Breitkopf SB, Cajka T, Cazenave-Gassiot A, Checa A, Cinel MA, Colas RA, Cremers S, Dennis EA, Evans JE, Fauland A, Fiehn O, Gardner MS, Garrett TJ, Gotlinger KH, Han J, Huang Y, Neo AH, Hyotylainen T, Izumi Y, Jiang H, Jiang H, Jiang J, Kachman M, Kiyonami R, Klavins K, Klose C, Kofeler HC, Kolmert J, Koal T, Koster G, Kuklenyik Z, Kurland IJ, Leadley M, Lin K, Maddipati KR, McDougall D, Meikle PJ, Mellett NA, Monnin C, Moseley MA, Nandakumar R, Oresic M, Patterson RE, Peake D, Pierce JS, Post M, Postle AD, Pugh R, Qui Y, Quehenberger O, Ramrup P, Rees J, Rembiesa B, Reynaud D, Roth MR, Sales S, Schuhmann K, Schwartzman ML, Serhan CN, Shevchenko A, Somerville SE, John-Williams LS, Surma MA, Takeda H, Thakare R, Thompson JW, Torta F, Triebl A, Trotzmuller M, Ubhayasekera SJK, Vuckovic D, Weir JM, Welti R, Wenk MR, Wheelock CE, Yao L, Yuan M, Zhao XH, Zhou S.
Harmonizing Lipidomics: NIST Interlaboratory Comparison Exercise for Lipidomics using Standard Reference Material 1950 Metabolites in Frozen Human Plasma.
J Lipid Res, 2017 Dec;58(12):2275-2288. doi: 10.1194/jlr.M079012. Epub 2017 Oct 6.
PMID: 28986437
Mohammed Y, Pan J, Zhang S, Han J, Borchers CH.
ExSTA: External Standard Addition Method for Accurate High-throughput Quantitation in Targeted Proteomics Experiments.
Proteomics Clin Appl, 2018 Mar;12(2):1600180. doi: 10.1002/prca.201600180. Epub 2017 Oct 25.
PMID: 28895300
Urao N, Mirza RE, Corbiere TF, Hollander Z, Borchers CH, Koh TJ.
Thrombospondin-1 and disease progression in dysferlinopathy.
Hum Mol Genet, 2017 Dec 15;26(24):4951-4960. doi: 10.1093/hmg/ddx378.
PMID: 29206970
Dilworth D, Upadhyay SK, Bonnafous P, Edoo AB, Bourbigot S, Pesek-Jardim F, Gudavicius G, Serpa JJ, Petrotchenko EV, Borchers CH, Nelson CJ, Mackereth CD.
The basic tilted helix bundle domain of the prolyl isomerase FKBP25 is a novel double-stranded RNA binding module.
Nucleic Acids Res, 2017 Nov 16;45(20):11989-12004. doi: 10.1093/nar/gkx852.
PMID: 29036638
Popp R, Li H, LeBlanc A, Mohammed Y, Aguilar-Mahecha A, Chambers AG, Lan C, Poetz O, Basik M, Batist G, Borchers CH.
Immuno-Matrix-Assisted Laser Desorption/Ionization Assays for Quantifying AKT1 and AKT2 in Breast and Colorectal Cancer Cell Lines and Tumors.
Anal Chem, 2017 Oct 3;89(19):10592-10600. doi: 10.1021/acs.analchem.7b02934. Epub 2017 Sep 20.
PMID: 28853539
Pedde RD, Li H, Borchers CH, Akbari M.
Microfluidic-Mass Spectrometry Interfaces for Translational Proteomics.
Trends Biotechnol, 2017 Oct;35(10):954-970. doi: 10.1016/j.tibtech.2017.06.006. Epub 2017 Jul 26.
PMID: 28755975
Richard VR, Domanski D, Percy AJ, Borchers CH.
An online 2D-reversed-phase – Reversed-phase chromatographic method for sensitive and robust plasma protein quantitation.
J Proteomics, 2017 Sep 25;168:28-36. doi: 10.1016/j.jprot.2017.07.018. Epub 2017 Jul 28.
PMID: 28757464
Peixoto RJM, Alves ES, Wang M, Ferreira RBR, Granato A, Han J, Gill H, Jacobson K, Lobo LA, Domingues RMCP, Borchers CH, Davies JE, Finlay BB, Antunes LCM.
Repression of Salmonella host cell invasion by aromatic small molecules from the human fecal metabolome.
Appl Environ Microbiol, 2017 Sep 15;83(19):e01148-17. doi: 10.1128/AEM.01148-17. Print 2017 Oct 1.
PMID: 28754707
Li H, Popp R, Frohlich B, Chen MX, Borchers CH.
Peptide and Protein Quantification Using Automated Immuno-MALDI (iMALDI).
J Vis Exp, 2017 Aug 18;(126):55933. doi: 10.3791/55933.
PMID: 28872133
Guarna MM, Hoover SE, Huxter E, Higo H, Moon KM, Domanski D, Bixby MEF, Melathopoulos AP, Ibrahim A, Peirson M, Desai S, Micholson D, White R, Borchers CH, Currie RW, Pernal SF, Foster LJ.
Peptide biomarkers used for the selective breeding of a complex polygenic trait in honeybees.
Sci Rep, 2017 Aug 21;7(1):8381. doi: 10.1038/s41598-017-08464-2.
PMID: 28827652
Larina IM, Percy AJ, Yang J, Borchers CH, M Nosovsky A, I Grigoriev A, N Nikolaev E.
Protein expression changes caused by spaceflight as measured for 18 Russian cosmonauts.
Sci Rep, 2017 Aug 15;7(1):8142. doi: 10.1038/s41598-017-08432-w.
PMID: 28811532
Brodie NI, Popov KI, Petrotchenko EV, Dokholyan NV, Borchers CH.
Solving protein structures using short-distance cross-linking constraints as a guide for discrete molecular dynamics simulations.
Science Advances, 2017 Jul 7;3(7):e1700479. doi: 10.1126/sciadv.1700479. eCollection 2017 Jul.
PMID: 28695211
LeBlanc A, Michaud SA, Percy AJ, Hardie DB, Yang J, Sinclair NJ, Proudfoot JI, Pistawka A, Smith DS, Borchers CH.
Multiplexed MRM-based Protein Quantitation Using Two Different Stable Isotope Labeled Peptides for Calibration.
J Proteome Res, 2017 Jul 7;16(7):2527-2536. doi: 10.1021/acs.jproteome.7b00094. Epub 2017 Jun 2.
PMID: 28516774
Wang X, Han J, Hardie DB, Yang J, Pan J, Borchers CH.
Metabolomic profiling of prostate cancer by matrix assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry imaging using Matrix Coating Assisted by an Electric Field (MCAEF).
Biochim Biophys Acta, 2017 Jul;1865(7):755-767. doi: 10.1016/j.bbapap.2016.12.012. Epub 2016 Dec 23.
PMID: 28017863
Mohammed Y, van Vlijmen BJ, Yang J, Percy AJ, Palmblad M, Borchers CH, Rosendaal FR.
Multiplexed targeted proteomic assay to assess coagulation factor concentrations and thrombosis-associated cancer.
Blood Advances, 2017 Jun 20;1(15):1080-1087. doi: 10.1182/bloodadvances.2017007955. eCollection 2017 Jun 27.
PMID: 29296750
Li H, Han J, Pan J, Liu T, Parker CE, Borchers CH.
Current Trends in Quantitative Proteomics — an update.
J Mass Spectrom, 2017 May;52(5):319-341. doi: 10.1002/jms.3932.
PMID: 28418607
Review
Qiu YL, Gong JY, Feng JY, Wang RX, Han J, Liu T, Lu Y, Li LT, Zhang MH, Sheps JA, Wang NL, Yan YY, Li JQ, Chen L, Borchers CH, Sipos B, Knisely AS, Ling V, Xing QH, Wang JS.
Defects in MYO5B are associated with a spectrum of previously undiagnosed low γ-glutamyltransferase cholestasis.
Hepatology, 2017 May;65(5):1655-1669. doi: 10.1002/hep.29020. Epub 2017 Mar 23.
PMID: 28027573
Reynolds LA, Redpath SA, Yurist-Doutsch S, Gill N, Brown EM, van der Heijden J, Brosschot TP, Han J, Marshall NC, Woodward SE, Valdez Y, Borchers CH, Perona-Wright G, Finlay BB.
Enteric helminths promote Salmonella co-infection by altering the intestinal metabolome.
J Infect Dis, 2017 Apr 15;215(8):1245-1254. doi: 10.1093/infdis/jix141.
PMID: 28368463
Streijger F, Skinnider M, Rogalski JC, Balshaw R, Shannon CP, Prudova A, Belanger LM, Ritchie L, Tsang A, Christie S, Parent S, Mac-Thiong JM, Bailey C, Urquhart J, Ailon T, Paquette SJ, Boyd MC, Street J, Fisher CG, Dvorak MF, Borchers CH, Foster LJ, Kwon BK.
A Targeted Proteomics Analysis of Cerebrospinal Fluid after Acute Human Spinal Cord Injury.
J Neurotrauma, 2017 Jun 15;34(12):2054-2068. doi:10.1089/neu.2016.4879. Epub 2017 Apr 7.
PMID: 28276985
Percy AJ, Michaud SA, Jardim A, Sinclair NJ, Zhang S, Mohammed Y, Palmer AL, Hardie DB, Yang J, LeBlanc AM, Borchers CH.
Multiplexed MRM-based assays for the quantitation of proteins in mouse plasma and heart tissue.
Proteomics, 2017 Apr;17(7). doi: 10.1002/pmic.201600097. Epub 2016 Nov 18.
PMID: 27688154
Mohammed Y, Bhowmick P, Smith DS, Domanski D, Jackson AM, Michaud SA, Malchow S, Percy AJ, Chambers AG, Palmer A, Zhang S, Sickmann A, Borchers CH.
PeptideTracker: A knowledgebase for collecting and storing information on protein concentrations in biological tissues.
Proteomics, 2017 Apr;17(7). doi: 10.1002/pmic.201600210. Epub 2016 Dec 14.
PMID: 27683069
Sobsey CA, Froehlich B, Batist G, Borchers CH.
Immuno-MALDI-MS for Accurate Quantitation of Targeted Peptides from Volume-Restricted Samples.
In: Methods in Molecular Biology, 2515: 203-225 (2022).
Percy AJ, Borchers CH.
Detailed Method for Performing the ExSTA Approach in Quantitative Bottom-up Plasma Proteomics.
In: Methods in Molecular Biology, 2228: 353-384 (2021).
Parker CE, Warren Hines MRE, Mocanu V, Greer SF, Borchers CH.
Mass Spectrometric Determination of Protein Ubiquitination.
In: Methods in Molecular Biology, 1934: 191-221 (2019).
Petrotchenko EV, Serpa JJ, Borchers CH
Cross-linking Applications in Structural Proteomics.
In: Proteomics for Biological Discovery, Wiley, 2019, 175-196, Veenstra TD and JR Yates (Eds.).
Eshghi A, Borchers CH.
Multiple Reaction Monitoring Using Double Isotopologue Peptide Standards 6 for Protein Quantification.
In: Methods in Molecular Biology, 1788: 193-214 (2018).
Percy AJ, Yang J, Chambers AG, Mohammed Y, Miliotis, Borchers CH.
Protocol for Standardizing High-to-Moderate Abundance Protein Biomarker Assessments Through an MRM-with-Standard-Peptides Quantitative Approach.
In: Modern Proteomics – Sample Preparation, Analysis and Practical Applications, Advances in Experimental Medicine and Biology, Springer,
2016, Vol. 919, 515-531
Percy AJ, Yang J, Chambers AG, Borchers CH.
Increased Depth and Breadth of Plasma Protein Quantitation via Two-Dimensional Liquid Chromatography/Multiple Reaction Monitoring-Mass Spectrometry with Labeled Peptide Standards.
In: Methods in Molecular Biology, 1410: 1-21 (2016).
Han J, Parker, CE, Borchers CH.
Chemical derivatization to enhance metabolomic analysis by LC/ESI–MS.
In: Advanced LC-MS Applications in Metabolomics, Future-Science eBook, Han J, Parker, CE, Borchers CH, Borchers CH (Eds), 2015.
Percy AJ, Chambers AG, Parker CE, Borchers CH.
MRM-based Protein Quantification with Labeled Standards for Biomarker Discovery, Verification, and Validation in Human Plasma.
In: Quantitative Proteomics, Royal Society of Chemistry, 2014, Vol. 1, 316-325.
Parker CE, Domanski D, Percy AJ, Chambers AG, Camenzind AG, Smith DS, Borchers CH.
Mass Spectrometry in High-Throughput Clinical Biomarker Assays: Multiple Reaction Monitoring.
In: Topics in Current Chemistry, Springer, 2014, Vol. 336, 117-138.
Petrotchenko EV, Makepeace KAT, Serpa JJ. Borchers CH.
Analysis of protein Structure by Crosslinking Combined with Mass Spectrometry.
In: Methods in Molecular Biology, Humana Press, 2014, Vol. 1156, 447-463.
Percy AJ, Chambers AG, Parker CE, Borchers CH.
A Targeted Mass Spectrometric Approach for Quantitating Candidate Cancer Biomarker Proteins in Undepleted and Non-Enriched Human Plasma.
In: The Research and Biology of Cancer, iConcept book, 2014, Vol. 1, 33-48.
Chen YT, Parker CE, Chen HW, Chen CL, Domanski D, Smith DS, Wu CC, Chung T, Liang KH, Chen MC, Chang YS, Borchers CH, Yu JS.
Discovery and Validation Case Studies, Recommendations: A Pipeline that Integrates the Discovery and Verification Studies of Urinary Protein Biomarkers Reveals Candidate Markers for Bladder Cancer.
In: Comprehensive Biomarker Discovery and Validation for Clinical Application, Royal Society of Chemistry, 2013, Vol. 33, Chapter 10, 271-314.
Shah B, Reid JD, Kuzyk MA, Parker CE, Borchers CH.
Developing an iMALDI Method.
In: Methods in Molecular Biology, 1023: 97-120 (2013).
Kuzyk MA, Parker CE, Domanki D, Borchers CH.
Development of MRM-based Assays in Absolute Quantitation of Plasma Proteins.
In: Methods in Molecular Biology, Humana Press, 2013, Vol. 1023, 53-82.
Percy AJ, Chambers AG, Parker CE, Borchers CH.
Absolute Quantitation of Proteins in Human Blood by Multiplexed Multiple Reactions Monitoring Mass Spectrometry.
In: Methods in Molecular Biology, Humana Press, 2013, Vol.1000, 167-189.
Percy AJ, Chambers AG, Parker CE, Borchers CH.
Absolute quantitation of proteins in human blood by multiplexed reaction monitoring mass spectrometry.
In: Methods in Molecular Biology, 1000: 167-89 (2013).
Domanski D, Smith DS, Miller CA, Yang Y, Jackson AM, Cohen Freue G, Hill JS, Parker CE, Borchers CH.
High-Flow Multiplexed MRM-Based Analysis of Proteins in Human Plasma Without Depletion or Enrichment.
In: Clinical Mass Spectrometry, Saunders, 2011, Vol. 31, N. 3, 371-384.
Shah B, Kozlowski RL, Han J, Borchers CH.
Emerging Mass Spectrometry-Based Technologies for Analyses of Chromatin Changes: Analysis of Histones and Histone Modifications.
In: Methods in Molecular Biology, vol. 773, Editor: Allison R. Kernmode (ed), Seed Dormancy: Methods and Protocols, 2011, Vol. 773, 259-303.
Ohlund LB, Hardie DB, Elliott MH, Smith DS, Reid JD, Cohen-Freue GV, Bergman AP, Sasaki M, Robertson L, Balshaw RF, Ng RT, Mui A, McManus, BM, Keown PA, McMaster WR, Parker CE, Borchers CH.
Standard Operating Procedures and Protocols for the Preparation and Analysis of Plasma Samples using the iTRAQ Methodology.
In: Sample Preparation in Biological Mass Spectrometry, Editor: Ivanov A, and Lazarev A, Springer, New York, 2011, 575-624.
Ouvry-Patat SA, Torres MP, Gelfand CA, Quek HH, Easterling M, Speir JP, Borchers CH.
Top-down proteomics on a high-field Fourier transform ion cyclotron resonance mass spectrometer.
In: Methods in Molecular Biology, 492: 215-31 (2009).
Petrotchenko EV, Borchers CH.
Cross-Linking as a Tool Examine Protein Complexes: Examples of Crosslinking Strategies and Computational Modeling.
In: Mass Spectrometry Analysis For Protein-Protein Interactions and Dynamics, Editor: Chance M, John Willey & Sons, Inc. 2008, Chapter 8, 157-168.
Parker CE, Warren MR, Mocanu V, Greer SF, Borchers CH.
Mass spectrometric determination of protein ubiquitination.
In: Methods Molecular Biology, 2008, Vol. 446, 109-30.
Parker CE, Smith D, Suckau D, Borchers CH.
Mass Spectrometry-based Tissue Imaging.
In: Advanced Imaging in Biology and Medicine, Editor: Sensen CH, Hallgrimsson W, Benedikt, Springer New York, 2008, 131-146.
Robinette D, Neamati N, Tomer KB, Borchers CH.
Photoaffinity Labeling Combined with Mass Spectrometric Approaches as a Tool for Structural Proteomics.
In: Expert Review in Proteomics, 3(4): 399-408 (2006).
Parker CE, Mocanu V, Warren MR, Greer SF, Borchers CH.
Mass Spectrometric Determination of Protein Ubiquitination.
In: Methods in Molecular Biology, Editor: Patterson WC, Humana Press, NJ, 2005, Vol. 103, 153-173.
Parker CE, Warren MR, Loiselle D, Dechiva NN, Scarlett CO, Borchers CH.
Identification of Components of Protein Complexes.
In: Methods in Molecular Biology, Editor: Patterson WC, Humana Press, NJ, 2005, Vol. 103, 117-151.
Borchers CH, Chen T, Neamati N.
Application of Proteomics in Basic Biological Sciences and Cancer.
In: Molecular Carcinogenesis and the Molecular Biology of Human Cancer, Editor: Warshawsky D, Landolph JR, CRC, 200, 263-288.
Borchers C.
Automation and integration strategies in high-throughput genomics; Editorial advisory Board Member Commentaries.
In: High-Throughput Genomics: Maximizing Efficiency and Productivity for Drug Discovery and Development, Editor SJ, Walker TD, Howard GA,
Hawkins KE, Vrana JA, D. Lockwood & M.A. Branca, Cambridge Healthtech Institute, 2001,118-119.
Przybylski M, Borchers CH, Jetschke M, Schuhmacher R, Fiedler W, Mak, Glocker MO.
Molecular Approaches for the Characterization of Protein Tertiary Structures by Selective Chemical Modification and Mass Spectrometric Peptide Mapping.
In: Peptides-Chemistry, Structure and Biology, Editor: Hodges RS, Smith JA, Escom Science Publ., Leiden, 1994, 254-256.
Congratulations Mina Farshadi! Our MSc student in Dr. Borchers’ lab has been presented the 2023 FeMS Empowerment award.
We are very proud to announce that our scientist, Deema Qasrawi will be giving a podium presentation at the upcoming MSACL 2023 13th Annual Conference & Exhibits in Monterey, CA. Her presentation is entitled “A simplified proteomics LC-MRM-MS assay for determination of ApoE genotypes in plasma samples”. Make sure to attend her oral presentation in April.
We are thrilled to reveal that our lab technician, Dennis Lee has been accepted for a poster presentation at this year’s upcoming MSACL 2023 13th Annual Conference & Exhibits in Monterey, CA. His presentation is entitled “LC-MS assay for antibiotics in intensive care unit clinical setting”. Be sure to stop by his poster presentation in April.
Dr. Christoph Borchers presented at the Agilent Technologies Breakfast Session at HUPO in Cancun. He spoke of new MRM approaches for faster, higher multiplex and sensitive protein quantitation from biological matrixes including blood and tissues.
Two of our graduate students, Connie Sobsey and Georgia Mitsa presented at HUPO in Cancun during the Clinical Proteomics/Precision Medicine session. Georgia’s presentation is entitled “Clinical cancer research on metastases from refractory colorectal cancer using a multi-omics approach to improve current precision oncology treatment”. They identified several new therapeutic vulnerabilities and putative biomarkers to better inform clinical decision making. Connie’s presentation is entitled “Targeted proteomics to verify the association between translational control pathways and capivasertib response in cancer cell lines and patient tumours”.
We welcome a new colleague, Yassene Mohammed to the Segal Cancer Proteomics Center.
New funding announcement!
11 research teams from Québec’s four institutions who received more than $4 million in major genomics funding were announced by Genome Québec in October. One of the selected teams is led by Dr. Christoph Borchers.
We are very happy to announce that our scientist, Deema Qasrawi will be giving an oral presentation at the upcoming MANA conference in Edmonton, AB. Her presentation is entitled “Multiplexed dilute-and-shoot LC-MRM-MS clinical assay for determination of the metanephrines and catecholamines in human urine”. Make sure to attend her oral presentation in September.
Congratulations to the Segal Cancer Proteomics Centre for the renewal of the CFI-MSI funding for The Metabolomics Innovation Centre!
Read the complete story to learn how Canada’s leading national research facilities received significant investments to drive research and innovation.
We are very excited to announce that our scientist, Deema Qasrawi will be giving an oral presentation at the upcoming 70th ASMS Conference on Mass Spectrometry and Allied Topics in Minneapolis, MN. Her presentation is entitled “Multiplexed dilute-and-shoot LC-MRM-MS clinical assay for determination of the metanephrines and catecholamines in human urine”. Make sure to attend her presentation during the conference in June.
Congratulations Vincent Lacasse! Our PhD student of Dr. Borchers’ Lab took home the Gairdner poster award at the CSHRF+Gairdner symposium!
Analytica 2022 in Munich is in full swing. Come and check out the Clinical Proteomics Session chaired by our own director Dr. Christoph Borchers. Wednesday, June 22 at 12:30 ICM/Hall 2.
We have exciting news to share! Dr. René Zahedi and Dr. Christoph Borchers have both been recently announced as the new co-editors for Expert Review of Proteomics.
Be sure to check out the International Conference on Multi-omics Technologies for Precision Medicine happening November 22-23, 2021. You’ll also want to attend the 10th Symposium on Structural Proteomics happening November 24-25, 2021!
Molecular and Cellular Proteomics recently highlighted our paper “Proteomic Portraits Reveal Evolutionarily Conserved and Divergent Responses to Spinal Cord Injury” on their webpage!
The last decade was an eventful one for proteomics as the field saw a number of advances ranging from new workflows and applications to improvements in instrumentation and bioinformatic innovations. To get a sense of the key developments, we asked leading researchers in the field for their picks of the most notable achievements in proteomics throughout the 2010s.
Megagrant project from Center for Computational and Data-Intensive Science and Engineering have received funding from Russian Ministry of Science and High Education: Dr. Christoph Hermann Borchers and his new laboratory project “Next-Generation Proteomics to Improve Personalized Medicine and Health Care”. (Laboratory for Mass-spectroscopy), (duration 2 years, 33 mln RUB/yr).
The UVic group has contributed 954 of the over 2350 targeted human and mouse MRM-based assays to the Assay Portal of the National Cancer Institute
Doctoral candidate Georgia Mitsa of Dr. Christoph Borchers’ proteomics lab received two awards from McGill University:
We welcome two new colleagues, Vanessa Gaspar working on an FRQS-funded project and Evgeniy Petrotchenko who is an expert in Structural Proteomics. Make sure to check out Part 1 of Agilent’s omics Webinar series on Jan 25th.
We are proud to be part of the Montreal Cancer Consortium (MCC) that has been recently funded by the Terry Fox Foundation and other co-funders. “Working together, the MCC researchers will also aim to better understand how various aspects of the immune system relate to acute leukemia and why therapy works for some patients but not others. They hope to identify new biomarkers and novel targets that will respond to immunotherapy treatments.”
We are very proud to announce that our scientist, Deema Qasrawi will be giving a podium presentation at the upcoming MSACL 2023 13th Annual Conference & Exhibits in Monterey, CA. Her presentation is entitled “A simplified proteomics LC-MRM-MS assay for determination of ApoE genotypes in plasma samples”. Make sure to attend her oral presentation in April.

We are thrilled to reveal that our lab technician, Dennis Lee has been accepted for a poster presentation at this year’s upcoming MSACL 2023 13th Annual Conference & Exhibits in Monterey, CA. His presentation is entitled “LC-MS assay for antibiotics in intensive care unit clinical setting”. Be sure to stop by his poster presentation in April.
We are thrilled to reveal that our Associate Director, Evgeniy Petrotchenko, will be presenting an in-person talk on Structural Proteomics entitled “Discovery of a novel ivacaftor drug-binding site on the CFTR protein using photo-affinity labelling combined with mass spectrometry” at this year’s ASMS Conference. Be sure to catch it virtually or in person in November 2021.
3755 Côte Ste-Catherine Road
Montreal, Quebec H3T 1E2
For inquiries regarding Analytical Mass Spectrometry – Proteomics and Metabolomics, please contact:
You have the power to make a difference! Your gift will support vital research at the Lady Davis Institute for Medical Research (LDI) that will translate into disease prevention, improved diagnoses, earlier detection, new and enhanced therapies, a better quality of life, wellness and hope for all of us.
Faire la différence, vous avez ce don! Votre contribution soutiendra la recherche essentielle menée à l’Institut Lady Davis qui permettra la prévention des maladies, des diagnostics plus précis, des dépistages plus rapides, des traitements innovants et plus performants, une meilleure qualité de vie, le bien-être et l’espoir pour nous tous.
Copyright © 2024 | Lady Davis Institute for Medical Research/Jewish General Hospital
Conception et développement : Yankee Media